Keywords: Sensitivity analysis
What data needs were addressed? To identify the most important parameters through sensitivity analysis of the IPCC Tier 2 model.
Why was the data needed? Having applied the IPCC Tier 2 method to country-specific data, researchers wanted to identify the most important factors driving emissions in order to prioritize future data improvements and research efforts so as to improve livestock GHG emission estimates and reduce the uncertainty of estimates for Senegal.
Methods used: Sensitivity analysis using regression methods.
How was the data need addressed? Senegal is a tropical country in West Africa, with an estimated cattle population of 3.4 million. Extensive livestock systems in Senegal are based on two main breeds of cattle: zebu Gobra (Bos indicus) in the North and taurine Ndama (Bos taurus) in the South. Together, these two breeds account for about 90% of the cattle population. To quantify emissions from these breeds using the IPCC Tier 2 model, a variety of data sources were used to derive input values. Information mainly came from two national livestock research centers (the Centre de Recherches Zootechniques de Dahra, CRZD and the Centre de Recherches Zootechniques de Kolda, CRZK, which are located in the sylvopastoral and agrosylvopastoral zones of Senegal, respectively). Both research centers frequently collect data through surveys and direct measurements on reproductive (e.g. fertility, calving) and productive (e.g. live weight, weight gain, milk yield) performance of cattle. Consequently, research reports, theses, publications and data from partnerships with international research organizations (e.g. FAO, ILRI, ITC) were used, together with documents from the Livestock Ministry of the Senegalese Government and Regional Centres on Agricultural Statistics. When local information was not available, expert judgement (e.g. for proportion of breeds in the cattle herd) or IPCC default values were used. Tables 1 and 2 show the input values used in the Tier 2 models for lactating cows and draft oxen.
Table 1: Assigned values of input parameters in the Tier 2 model for Gobra and Ndama lactating cows
| Parameter | Symbol | Unit | Used value | |
|---|---|---|---|---|
| Gobra | Ndama | |||
| Average daily weight gain | ADG | kg/day | 0.135 | 0.110 |
| Coefficient | C | dimensionless | 0.8 | 0.8 |
| Activity coefficient | Ca | MJ/day/kg | 0.36 | 0.36 |
| Maintenance coefficient | Cfi | MJ/day/kg | 0.386 | 0.386 |
| Pregnancy | Cp | dimensionless | 0.10 | 0.10 |
| Feed digestibility | DE | % | 50 | 50 |
| Fat content of milk | Fat | % | 4.7 | 4.24 |
| Average life body weight | LW | kg | 250 | 200 |
| Milk yield | Milk | kg/day | 0.922 | 0.870 |
| Mature life body weight | MW | kg | 200 | 180 |
| Methane conversion rate | Ym | % | 6.5 | 6.5 |
Table 2: Assigned values of input parameters in the Tier 2 model for Gobra and Ndama draft ox
| Parameter | Symbol | Unit | Used value | |
|---|---|---|---|---|
| Gobra | Ndama | |||
| Average daily weight gain | ADG | kg/day | 0.135 | 0.110 |
| Coefficient | C | dimensionless | 1.2 | 1.2 |
| Activity coefficient | Ca | MJ/day/kg | 0.36 | 0.36 |
| Maintenance coefficient | Cfi | MJ/day/kg | 0.37 | 0.37 |
| Feed digestibility | DE | % | 50 | 50 |
| Average amount of work | Hour | h/day | 1.23 | 1.23 |
| Average life body weight | LW | kg | 300 | 250 |
| Mature life body weight | MW | kg | 200 | 180 |
| Methane conversion rate | Ym | % | 6.5 | 6.5 |
The purpose of conducting sensitivity analysis was to identify which parameters used in the development of methane enteric emission factor require additional research in order to reduce output uncertainty. To do this, the ‘sensitivity’ package (Pujol et al. 2012) implemented in R software (version 3.3.3) was used. First, we defined the possible ranges of values for each parameter and values were generated between the minimum and the maximum of each parameter used in the sensitivity analysis. For all parameters (e.g. milk, liveweight), we assumed a uniform distribution (with a 95% confidence interval) of ± 20% around each used value. These values were input into the IPCC model to produce a range of values for the output (i.e. annual methane enteric emissions per head). Finally, a regression technique was performed to obtain sensitivity indices (i.e., standardized regression coefficient) for each parameter in the model.
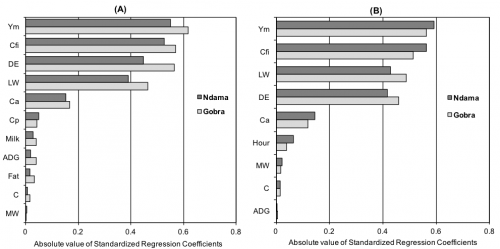
The linear regression method shows sensitivity indices for each input parameter used to estimate enteric methane EF (Figure 1). Overall, the results reveal that for lactating cows and draft oxen the methane conversion rate (Ym), the coefficient for calculating net energy for maintenance (Cfi), digestible energy (DE) and liveweight (LW) are the most important parameters affecting the estimated emission factors. Thus, future research should prioritize producing improved estimates of these parameters. While there is relatively more information on live weight and feed digestibility in the Sub-Saharan Africa region, very little research has been conducted on methane conversion rates or other coefficients in the IPCC model. Direct measurements of methane output per unit of feed intake using SF6 tracer techniques or respiration chambers would be necessary to improve estimates of cattle methane emissions in Senegal.
Figure 1: Standardized regression coefficients of input parameters used to calculate enteric methane emission factors for lactating cows (figure A) and draft oxen (figure B) of Gobra and Ndama cattle
Further Resources
Faivre R, et al. 2013. Analyse de sensibilité et exploration de modèles application aux sciences de la nature et de l’environnement. Editions Quae.
Hamby DM. 1994. A review of techniques for parameter sensitivity analysis of environmental models. Environmental monitoring and assessment.
Iooss B. 2011. Revue sur l’analyse de sensibilité globale de modèles numériques. Journal de la Société Française de Statistique.
Makowski D, et al. 2006. Global sensitivity analysis for calculating the contribution of genetic parameters to the variance of crop model prediction. Reliability Engineering & System Safety.
Pujol G, et al. 2012. The R package “sensitivity”, version 1.6-1. CRAN, Technical report.
Saltelli A. 2002. Sensitivity analysis for importance assessment. Risk analysis.
This inventory practice note was contributed by Séga Ndao, El Hadji Traore and Mamadou Diop. For further information, contact ndaosega@gmail.com.

